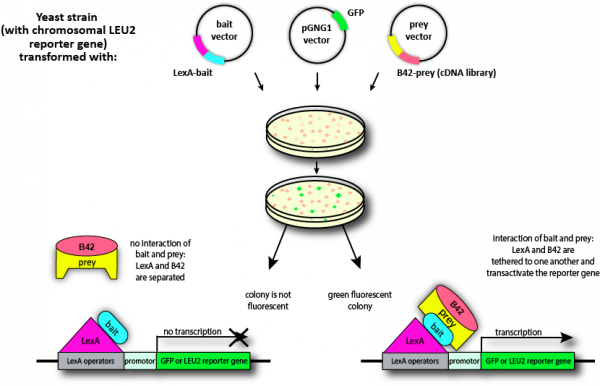
| Description: | Grow'n'Glow GFP Yeast Two-Hybrid System. The two-hybrid assay provides a sophisticated and elegant way to investigate protein interactions. In addition to being able to characterize the domains, amino acid residues, and conditions necessary for known protein interactions, new interacting proteins can be identified. This screening ability makes the two-hybrid approach a powerful tool for gene discovery and functional analysis. The Grow'n'Glow GFP system simplifies and accelerates the screening process of two-hybrid systems. It combines the advantages of the well-known gene expression reporter protein GFP (green fluorescent protein) with the LexA based yeast two-hybrid technology. As a result, protein-protein interactions can be detected in vivo in a one-step selection under UV light. Interacting proteins result in expression of two reporter genes - the LEU2 nutritional selection gene and the GFP gene from Aequorea victoriae. Expression of LEU2 allows yeast with interacting proteins to grow on medium lacking leucine, and expression of the GFP gene is readily detected by visualizing colonies under ultraviolet (UV) light. Use of the tightly controlled LEU2 and the easily detectable GFP reporters reduces background and offers more clear-cut results, which expedite the entire screening process. Our GFP two-hybrid system uses the E. coli LexA protein as a DNA binding protein to provide promoter specificity. Since LexA is a prokaryotic protein, there is little chance of getting non-specific activation, as it is the case when the GAL4 DNA-binding domain is used (since GAL4 is an endogenous yeast protein, it may interact with a variety of elements in a yeast cell). Use of the Green Fluorescent Protein (GFP) reporter eliminates the time-consuming beta-galactosidase lift assay. The particular GFP variant expressed by the pGNG1 plasmid, GFPuv, has the same excitation and emission maxima as wild-type GFP but is 18 times brighter than the wild-type variant. Yeast colonies with interacting proteins are glowing brightly green which can be easily detected by placing the plate with the yeast colonies on a standard 300 nm UV-table or under a UV-handlamp in a darkroom. Expression of fusion proteins by the prey plasmid is controlled by the GAL1 upstream-activation sequence. In yeast with an intact galactose regulatory system, the GAL1 activation sequence is induced by galactose and repressed by glucose. This regulation inhibits expression of library/activation domain fusions until the actual screening, which prevents any potentially toxic library proteins affecting growth of the yeast. This sort of toxic proteins would be missed during screening in a non-inducible system. Features: • Identifies in vivo protein-protein interactions of a bait protein of choice and proteins expressed from a cDNA library • Determines the protein binding regions of two proteins known to interact • Positive colonies fluoresce brightly green - therefore, one-step selection of potential positives under UV light • No time-consuming beta-galactosidase lift assays as essential in most conventional two-hybrid assays • Screens for potentially toxic interacting proteins using galactose-inducible expression of libraries Fewer false positives |
| Order #: | GNGK02 |
| Unit Size: | Kit |
| Supplier: | MoBiTec GmbH |
| Shipping: | RT |
| Storage: | 4 °C |
| Subcategory: | Two- and One-Hybrid Systems |
Add to request list
- Phone: +49 (0)89 3799666-6
- E-mail: info@biozol.de
MoBiTec is now part of BIOZOL
All orders and requests placed via the online shop on www.mobitec.com are processed by BIOZOL Diagnostica Vertrieb GmbH.
You will receive an order confirmation by fax or e-mail. More information regarding the ordering process can be found here.
Technical Service - Product Information
Viewed